10.2.1 Calculate and predict the genotypic and phenotypic ratio of offspring of dihybrid crosses involving unlinked autosomal genes
A dihybrid cross determines the allele combinations of offspring for two particular genes that are unlinked (not on the same chromosomes)
Because there are two genes with two alleles per gene (multiple alleles not required), there can be up to four different gamete combinations
10.2.2 Distinguish between autosomes and sex chromosomes
Autosomes: Pairs of chromosomes that are identical in appearance (e.g. same size, same gene loci, etc) and are not involved in sex determination
Sex chromosome: Pairs of chromosomes involved in sex determination and are not identical in appearance (e.g. X and Y chromosome in humans)
10.2.3 Explain how crossing over between non-sister chromatids of a homologous pair in prophase I can result in an exchange in alleles
During crossing over in prophase I, non-sister chromatids of a homologous pair may break and reform at points of attachment called chiasmata
As these chromatids break at the same point, any gene loci below the point of the break will be exchanged as a result of recombination
This means that maternal and paternal alleles may be exchanged between the maternal and paternal chromosomes, creating new gene combinations
The further apart two gene loci are on a chromosome, the more likely they are to be exchanged
10.2.4 Define linkage group
A linkage group is a group of genes whose loci are on the same chromosome and therefore do not follow the law of independent assortment
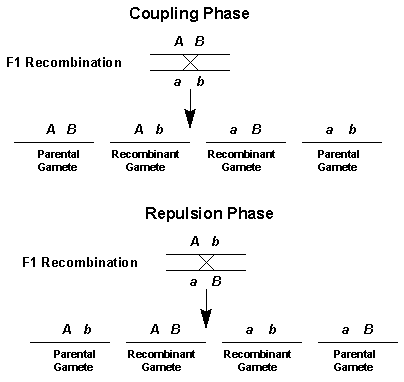
Linkage genes will tend to be inherited together - the only way to separate them is through recombination (via crossing over during synapsis)
10.2.5 Explain an example of a cross linkage between two linked genes
When two genes are linked, they do not follow the expected phenotypic ratio for a dihybrid cross between heterozygous parents
Instead the phenotypic ratio will follow that of a monohybrid cross as the two genes are inherited together
This means that offspring will tend to produce the parental phenotypes
Recombinant phenotypes will only be evident if crossing over occurs in prophase I and would thus be expected to appear in low number (if at all)
An example of a cross between two linked genes is the mating of a grey bodied, normal wing fruit fly with a black bodied, vestigial wing mutant
10.2.6 Identify which of the offspring are recombinants in a dihybrid cross involving linked genes
Recombinants of linked genes are those combinations of genes not found in parents
For example, in a test cross of a heterozygous fruit fly (grey bodied, normal wings) with a homozygous recessive mutant (black bodied, vestigial wings), the recombinants would be the grey bodied, vestigial winged offsprings and the black bodied, normal winged offspring
Linked genes that have undergone recombination can be distinguished from unlinked genes via test cross because the frequency of the recombinant genotype will always be less than would occur for unlinked genes (crossing over does not happen every time)
- For example:
- Heterozygous test cross of unlinked genes = 1:1:1:1 phenotypic ratio
- Heterozygous test cross of linked genes = 1:1:0.1:0.1 pehnotypic ratio (uncommon phenotypes are recombinants)